[ad_1]
Discussion
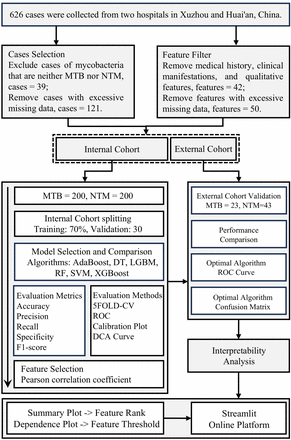
In this study, we conducted a retrospective analysis of the clinical characteristics of patients infected with MTB and NTM and developed six machine-learning models. Multiple evaluation methods and metrics demonstrated that the RF algorithm exhibited strong discriminative power and clinical utility. Subsequently, we applied Pearson correlation coefficients to eliminate multicollinearity among features and assessed the performance of the RF model in external validation. In addition, we employed SHAP to visualise the predictive model, facilitating the interpretation of key features and their threshold values in the model’s decision-making process. Finally, the constructed RF model was deployed to the cloud, and a user-friendly decision-support system was developed.
Current diagnostic approaches for TB primarily rely on microbiological methods, such as sputum smear microscopy and culture of mycobacteria on liquid or solid media.17 While smear microscopy is rapid and cost-effective, its sensitivity is limited, particularly in cases without cavitary lesions. Culture, regarded as the diagnostic gold standard due to its higher sensitivity, typically requires 2–6 weeks to yield results.5 Several omics-based approaches have identified novel biomarkers for distinguishing MTB and NTM. Pathway analyses of the proteomic and lipidomic profiles revealed significant regulation of the TB pathway, sphingolipid signalling and adiponectin signalling. Integration of immune-related protein and lipid biomarkers markedly enhanced the diagnostic specificity and sensitivity in distinguishing NTM from MTB.18 Targeted metabolomics further identified specific metabolites, such as meso-hydroxyheme and itaconate anhydride, that robustly discriminated between pre-extensively and extensively drug-resistant TB strains,19 yet these biomarkers lack rigorous and reliable external validation. ML approaches that integrate epidemiological features, risk factors and laboratory diagnostic parameters have also been employed to transform complex datasets into actionable diagnostic models. For instance, Takeuchi et al leveraged immune cell profiles, inflammatory mediators, chemokines and tissue injury to guide the management and treatment of bacterial infections in peritoneal dialysis patients.20 However, the mode’s applicability is constrained by the need for comprehensive and high-resolution clinical data, limiting its implementation to well-equipped hospital settings. In contrast, models developed using routine laboratory parameters offer several practical advantages. In this study, we highlight the potential of routine laboratory parameters in differentiating between MTB and NTM disease. A total of 49 clinical features from 466 patients were collected to construct ML models. Among various evaluation metrics, the RF algorithm demonstrated the best performance, achieving accuracies of 82.71% and 87.69% in internal and external validations, respectively, with robust sensitivity and specificity. Given that an excessive number of features may hinder its clinical applicability and compromise its accuracy,21 we conducted a Pearson correlation analysis to identify highly correlated features. The results showed that setting the correlation coefficient threshold to 0.75 yielded the best performance for the RF model, achieving an accuracy of 85.57%.
However, the acceptability of clinical diagnostic models by both clinicians and patients, as well as their integration into shared decision-making processes, constitutes a fundamental prerequisite for translating such models into clinical practice.22 We used DCA to evaluate the clinical utility of all models. Among them, the optimal diagnostic model, RF, demonstrated a net benefit in identifying patients requiring treatment for either MTB or NTM disease within a probability threshold range of 0.51–0.9. This finding suggests that, in scenarios where minimising missed NTM disease is a priority, a lower threshold may be preferred. To enhance clinicians’ trust in ML-based decisions and improve model transparency, the SHAP method was employed to provide both global and local interpretability.23 Contribution analyses for each feature in the model revealed findings that align with previous reports. Prior research, accounting for environmental, geographical and lifestyle factors, identified gender as the only retained variable in statistical models, with female patients exhibiting a higher susceptibility to NTM infection.24 Regarding HDL, patients with NTM pulmonary disease presented higher HDL levels relative to those with drug-resistant NTM.25 The procalcitonin biomarker demonstrated high specificity before and after MTB treatment.26 For ALB, evidence indicates that patients with NTM infection often display compromised nutritional status, with levels frequently falling below the normal range (40.00–55.00 g/L); in this study, the identified ALB cut-off was <39.00 g/L, closely matching clinical observations.27 Intriguingly, Cl and Na emerged as the two most important predictors in this study. While there is currently no prior evidence supporting these variables as independent discriminators between MTB and NTM, variations in their measured values appear to be associated with infection by either pathogen. Ultimately, to maximise generalisability and facilitate the translation of this research into real-world clinical practice, we developed a free webpage tool for clinicians and patients. Users can input the 10 real indicators to calculate the predicted probability of MTB or NTM.
Several limitations remain to be addressed. First, this study was conducted at only two hospitals with a relatively small sample size, lacking comprehensive multicentre validation and potentially introducing selection bias. Robust cross-validation and external validation indicate that future studies would benefit from multicentre, large-sample, randomised controlled trials to provide sufficient statistical power for the development of models with high accuracy and strong generalisability. Second, clinical data on immunodeficiency, such as HIV infection, organ transplantation or immunosuppressive therapy, were not available. As immunosuppression is a known risk factor for NTM disease, this may have introduced unmeasured confounding and limited the generalisability of the model across clinical populations. Third, all variables in this study were derived from routine laboratory tests, while many emerging biomarkers with promising diagnostic potential warrant inclusion in future models to enhance diagnostic performance. Fourth, samples with negative results on PCR-reverse dot blot hybridisation or smear microscopy were not included in model development, which limits the applicability of the model to patients with confirmed mycobacterial disease. Moreover, this study focused solely on distinguishing between MTB and NTM, without further subclassification of specific species such as Mycobacterium avium complex and Mycobacterium kansasii, and was also unable to differentiate between infected and non-infected individuals or identify cases of NTM–MTB coinfection. With continued experimental efforts and data accumulation, a more comprehensive diagnostic model for TB could be developed.
[ad_2]
Source link