[ad_1]
Discussion
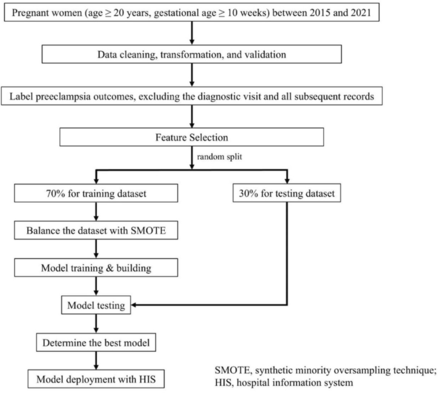
This study developed an ML model for predicting pre-eclampsia using routinely collected clinical data, achieving excellent discrimination (AUC 0.942). The model enables clinicians to provide patients with quantitative risk estimates, supporting earlier preventive interventions and individualised monitoring throughout pregnancy.
A major advantage of this approach is its reliance on variables readily available from electronic medical records, without the need for costly biomarkers or specialised ultrasound. The model can generate predictions from the tenth week of gestation and update risk dynamically as clinical parameters change, facilitating timely measures such as low-dose aspirin or intensified surveillance. Moreover, integration into the HIS allows seamless use during routine consultations, with outputs that are easy to interpret and communicate to patients. This feature enhances shared decision-making and empowers women to participate actively in their care. By accurately identifying both high-risk and low-risk patients, the tool has the potential to optimise resource utilisation, reduce unnecessary hospitalisations and improve overall pregnancy outcomes. As summarised in table 3, our study differs from previous work by covering all occurrences of pre-eclampsia rather than restricting to early onset or late onset.13 14 Unlike Li et al27 who examined intrapartum and postpartum cases, our model predicts risk as early as the tenth week and continues to update throughout pregnancy. Unlike prior studies using multi-institutional databases with variable diagnostic criteria,18 27 28 our single-centre design ensured consistent standards, which likely contributed to the higher AUC. This uniformity in data collection is a key strength, yielding a reliable data set and supporting the superior performance of our model based on readily available clinical variables. Consistent with prior literature, our study confirmed significant associations between pre-eclampsia and patients’ comorbidities, as well as routine measures such as HbA1c. SHAP analysis identified DBP as the strongest predictor, followed by SBP and urine glucose. Clinically, this highlights the need to monitor DBP with equal attention as SBP and to consider urine glucose in routine assessments, which may further improve early identification and management of high-risk patients. Consequently, following the completion of the medical consultation, this AI prediction model can offer personalised outcome predictions. These predictions are provided in real time and are easily comprehensible for both patients and clinicians, with quantifiable data. By using this prediction model, clinicians and patients can make more confident decisions regarding subsequent treatment strategies. Importantly, the clinical interpretation of a high predicted risk at a given prenatal visit warrants clarification. Because our model was designed to generate a risk probability at each antenatal encounter prior to the actual diagnosis of pre-eclampsia, a high score should be viewed not as a marker of imminent diagnosis at the next visit, but rather as an indication that the woman is at a substantially elevated likelihood of developing pre-eclampsia later in pregnancy. In practice, this per-visit risk stratification can serve two complementary purposes: first, to flag women who may benefit from intensified monitoring in the short term (eg, closer follow-up, repeat laboratory testing); and second, to inform longer-term management plans such as prophylactic aspirin, increased surveillance in the third trimester or early delivery planning if clinically indicated. This dual interpretation underscores that the model is not intended to replace clinical judgement, but to provide dynamic, evidence-based risk estimates that support timely intervention across different stages of pregnancy. Despite its strengths, there were several limitations in this study. First, the limited sample size and shortage of input data may have adversely impacted the model’s performance. Additionally, the study was conducted using data from a single hospital, which may limit the generalisability of the findings to broader populations. However, we believe that the consistent diagnostic criteria used within the hospital helped strengthen the reliability of the data. Moreover, Taiwan, being a relatively small region with an integrated healthcare system, has generally comparable patient populations across different hospitals. Nonetheless, future studies should aim to include multicentre data to enhance the external validity of the findings. Furthermore, because diagnostic criteria and antenatal care schedules may vary across institutions, the outcome labelling rule and per-visit prediction framework established in this study may require adaptation before being applied in other clinical settings. This underscores the importance of external validation to confirm the broader applicability of our approach. Second, we did not assess other laboratory feature variables such as pregnancy-associated plasma protein-A, soluble Fms-like tyrosine kinase-1 and placenta growth factor. Including these variables could potentially improve the accuracy of the results. However, since the aim of this study was to develop a prediction model that could be implemented solely using electronic medical record data, we opted not to include these expensive and less accessible laboratory tests. Third, the patient information we collected did not differentiate whether aspirin was used, which could potentially affect the predictive outcomes and subsequent effectiveness of using medication for prevention. Despite these limitations, we believe that establishing an ML model and integrating it with the HIS can assist clinicians in predicting patient risks and developing personalised treatment plans. To enhance the robustness and predictive accuracy of the model, we recommend applying various balancing techniques, including SMOTE, to mitigate the risks of model bias and overfitting. In addition, although all available variables were included to maximise predictive accuracy, future studies may benefit from applying systematic feature selection techniques. Such approaches could reduce model complexity, enhance interpretability and further improve predictive performance. While this study focused on clinical and personal history data, future studies could benefit from incorporating molecular-level data, such as transcriptomics or proteomics, to capture the heterogeneity and complexity of pre-eclampsia.29 30 Integrating such data with ML models may improve the accuracy and early prediction of pre-eclampsia, especially in more complex cases.
[ad_2]
Source link