Methods
This is a retrospective diagnostic test accuracy study. The study methodology and reporting follow the recommendations of Standards for Reporting Diagnostic Accuracy (STARD).11 The STARD checklist is included in the online supplemental file 1.
Design, setting, population and data source
We assessed the performance of three methods of CT report digitalisation: an investigator-programmed text classification (TC) programme (in R: a language and environment for statistical computing R Foundation for statistical computing, Vienna, Austria), a commercially available clinical NLP approach (CLiX, Clinithink, Alpharetta, Georgia, www.clinithink.com)12 and an investigator-designed large language model (LLM) approach using generative pretrained transformer (GPT).
This is a subgroup analysis of an ongoing study based in the UK. Institutional ethics approval was obtained from the Health Research Authority and Health and Care Wales (research ethics committee reference 24/LO/0103).
Patients presenting to any of the three EDs of Barts Health NHS Trust, London, UK, between 1 January 2018 and 31 December 2022 with a coded chief complaint of head injury (SNOMED-CT 170720001) and who had a CT scan of the head performed were eligible. Age, sex, the narrative CT report and other clinical and electronic data were extracted from the electronic patient record system. A random sample of 900 cases was selected from the overall study population to serve as the data source for this study.
Definitions, data handling and generation of gold-standard interpretation
ICB was defined as present (ICB+) if there was bleeding in any intracranial space (extradural, subdural, subarachnoid, intraventricular or intraparenchymal, including contusion). Head CTs were classified as ICB− if there were no haemorrhages or if there was bleeding limited to extracranial anatomical regions (eg, subgaleal, facial).
The reference standard classification for each CT report was determined by manual classification by research physicians. Two physicians reviewed the reports (SL and SHT). For 10% of CT reports, a second researcher (BB), blinded to the results of the initial review, provided an additional evaluation. Any classification uncertainties by the initial two researchers were adjudicated by a senior author (BB). All adjudicators of the reference standard were blinded to the results of the three digitisation methods.
Manual ICB classifications were entered into a spreadsheet (Excel, Office365, microsoft.com). Results from the tested digitalisation methods were added into the spreadsheet before importing data into analysis software (Stata 18.5MP, Stata Corp, stata.com).
Digitalisation procedures and reporting
Three digitalisation methods were assessed in this project. The first was a text classifier (TC) developed in R. This was created using regular expressions to extract relevant vocabulary, Doc2Vec to vectorise the expressions, and a random forest model as a classifier for whether each sentence with relevant vocabulary affirmed or rejected ICB+. The second was a proprietary artificial intelligence (AI) information extraction (Clinithink, www.clinithink.com). The third was an investigator-programmed approach using LLM. Basic characteristics of the assessed digitalisation methods are provided in table 1, with further details (including programming information) outlined in sections S2-1, S2-2 and S2-3 of the online supplemental file 1.
•
Three digitalisation approaches assessed in this study
The TC and Clinithink methods allowed only for dichotomous categorisation (ICB+ or ICB−). Developing the TC approach required a derivation set, which was constituted by randomly selecting half of the overall dataset. The decision to split the data into two equal sets for derivation and validation was made a priori and planned based on predicted processes in which regular expressions were refined (online supplemental section S2-1). The vocabulary was refined on half of the available reports, with the aim of focussing on more commonly used expressions and avoiding those used rarely (once or twice). This split was then carried into the remaining train/testing. The derivation set (n=449) was used for TC development and optimisation. The remaining 449 head CT reports constituted the ‘test set’ used for TC validation.
Clinithink does not require a training set for development. Clinithink provides explainability of where in the document and which postcoordinated SNOMED-CT terms were used to classify a document as ICB+/ICB−.
For both the TC and Clinithink, the reporting of results in terms of probability of ICB, or P(ICB), assumed values of either 0 or 1. The main evaluation of the commercial method was executed on the same 449 cases used for R validation.
To develop DECIPHER-LLM, a total of 50 samples from the 898 were used in prompt engineering and optimisation via the GPT-4o-mini Application Programming Interface (API). 50 samples were selected to ensure a diversity of cases for the prompt engineering process while avoiding potential overfitting. DECIPHER-LLM was designed to report a probability of ICH, rather than dichotomous output, as well as to offer reasoning as to why the algorithm applied that probability. We used Liu and Youden methods to select a probability threshold above which a CT was interpreted as having ICB+.13 We then replicated the analyses performed by TC and Clinithink, wherein DECIPHER-LLM was evaluated against the same dataset (n=449).
In a secondary analysis, we combined DECIPHER-LLM with manual review in a ‘hybrid’ model. For this analysis, we selected a probability threshold that maintained a sensitivity of 100% as our cut-off. We then calculated sensitivity, specificity and number needed to evaluate for this cut-off.
The main comparisons between the three digitalisation methods used the same 449-record test set. Since no derivation set was required for Clinithink or DECIPHER-LLM, the derivation dataset (also of n=449) that had been used to develop the TC was used as a supplemental (internal) evaluation dataset for the remaining two methods. This served as a planned sensitivity analysis to ascertain whether the performance of the Clinithink and DECIPHER-LLM classification methods substantively changed when applied to a different dataset.
Analysis methods
Stata was used for all analysis and plotting. Significance was defined at the p<0.05 level, and CIs were calculated at the 95% level. Non-overlap of CIs was interpreted as suggesting statistical significance at the p<0.05 level.
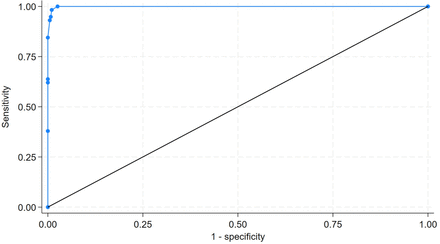
True and false positives (TP and FP) and true and false negatives (TN and FN) were assessed. We calculated standard performance measures: sensitivity, specificity, accuracy, positive and negative predictive values (PPV and NPV) and likelihood ratios associated with positive (LR+) and negative (LR−) classification. Classification performance was assessed using the Matthews correlation coefficient (MCC) and the F1 score.14 Model calibration was assessed using Brier Scores. Area under the receiver operator characteristic curve (AUROC) was calculated. AUROC comparisons were performed using a χ2 comparator, which incorporates Sidak’s multiple comparison correction.
Because from both clinical and research perspectives, minimising false negatives is critical, we prioritised sensitivity. A cut-point was set for the DECIPHER-LLM, at varying levels of LLM-calculated probability of positive testing P(ICB). The concept of the cut-point was based on the fact that DECIPHER-LLM could be set to report not just a ‘yes or no’ for predicting ICB but also a P(ICB) between 0% and 100%. The cut-point could be set at varying levels of P(ICB), to identify the lowest cut-point for P(ICB) that maintained a sensitivity of 100%. We therefore identified the maximum DECIPHER-LLM P(ICB) that retained 100% sensitivity and then obtained the count of CT reports that could be eliminated from the manual review requirement. The proportion of these reports (ie, those reliably classifiable as ICB−) was calculated and reported with a 95% CI.