[ad_1]
Discussion
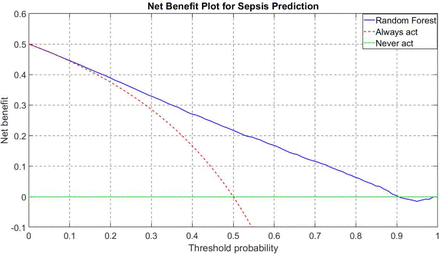
The results obtained in this study highlight the potential of AI for early detection of sepsis in the ED using free text analysis of patient concerns prior to clinical assessment. The random forest classifier demonstrated the best predictive performance among the models evaluated, with an AUPRC of 0.7890 and an AUC of 0.8000. Notably, the BERT model, which leverages advanced NLP techniques to process raw text data, also achieved high predictive performance, with an AUPRC of 0.7542 and an AUC of 0.7735. While slightly lower than the random forest model, BERT’s strong performance highlights the utility of leveraging unstructured triage narratives for sepsis prediction. The fact that both models performed well supports the feasibility of incorporating free-text data into AI-driven clinical decision support tools at ED triage, where early detection is critical for improving patient outcomes. The elastic-net feature selection algorithm identified nine key features across all folds, including demographic factors (age), clinical indicators (pain, temperature) and process metrics (ED treat to discharge time). These features provide valuable insights into the predictive factors associated with sepsis development.
Our findings align with previous research by Horng et al,19 which demonstrated the effectiveness of incorporating free-text data into AI-based clinical decision support systems for sepsis prediction in the ED. Their retrospective study (2008–2013) used ICD-9-CM discharge diagnoses and divided patients into training (64%), validation (20%) and test (16%) groups. They created four models integrating vital signs, chief concerns and preprocessed free text, using bag-of-words and topic models with a support vector machine for prediction. Out of 230 936 patient visits, about 14% were diagnosed with infection. The best-performing models, which included free text data, achieved an AUC of 0.86 in the test set.
In addition to establishing an AI model to detect sepsis, we were also interested in the prevalence of sepsis in a large ED. One of the difficulties for clinicians is maintaining vigilance across the many ‘common’ conditions that present to the ED. Initially, 5.9% of patients in the data set had sepsis ICD codes assigned during their admission. After excluding patients with potential HACs, the prevalence reduced to 4.1%. Given the possibility that some of the removed HAC cases might still be relevant sepsis cases, we estimate the prevalence of ‘real’ sepsis to be between 4.1% and 5.9% in our data set.
Our results also align with those reported by the Australian Institute of Health and Welfare, which indicated a 4.9% prevalence of ‘Certain infectious and parasitic diseases’ in ED presentations in Western Australia.22 Sepsis accounts for 6.7% of healthcare expenditure in the region, highlighting its substantial economic burden. Our findings support using free-text data in ML models to enhance sepsis diagnosis in ED patients. By analysing clinical notes and narrative descriptions, these models can identify sepsis cases that may be missed when relying only on structured data like vital signs and demographics. Our study, using limited ED data and triage comments, demonstrates the potential of ML, particularly random forest, in early sepsis detection. These models, leveraging demographic, clinical and temporal features, can help clinicians identify at-risk patients promptly, leading to timely interventions and improved outcomes. Identified features such as pain levels, vital signs and treatment duration emphasise the importance of comprehensive patient assessment in identifying sepsis risk. Integrating these predictors into clinical decision support systems can enhance diagnostic accuracy and facilitate targeted interventions, reducing morbidity and mortality associated with sepsis.
Automating decision support for sepsis detection at ED triage is challenging,23 24 as previous studies25 26 often rely on lab results and continuous vital sign monitoring, which are not always available in fast-paced ED settings. Thus, clinical practice continues to use vital signs as a trigger for diagnosis of sepsis at triage.27 28 Consequently, vital signs alone are still used but are neither sensitive nor specific enough for early sepsis detection. Leveraging all available data, including free text from clinical notes, can improve decision support triggers, and our results, supported by previous studies,19 highlight this opportunity.1
Previous systematic reviews and meta-analyses on AI for sepsis prediction highlight its effectiveness.13 18 Kijpaisalratana et al29 used retrospective data from adult ED patients, and used algorithms including logistic regression, gradient boosting, random forest and neural networks. Their models, trained on 80% of the data and tested on 20%, outperformed traditional models including quick Sequential Organ Failure Assessment, Modified Early Warning Score and systemic inflammatory response syndrome with a higher area under receiver operating characteristic curve of 0.93 for the best model (random forest). These findings support the superiority of ML over traditional methods for predicting sepsis in ED patients.
Fleuren et al18 conducted a systematic review and meta-analysis of 28 studies, evaluating 130 ML models for real-time sepsis diagnosis. They reported that the diagnostic accuracy (measured by AUC) varied by setting: 0.68–0.99 in the ICU, 0.96–0.98 in-hospital and 0.87–0.97 in the ED. Despite varying sepsis definitions, the models accurately predicted sepsis, highlighting the need for further research to bridge the gap between data and clinical practice.
[ad_2]
Source link