Abstract
Objective Data-driven innovations are essential in strengthening disease control. We developed a low-cost, open-source system for robust epidemiological intelligence in response to the COVID-19 crisis, prioritising scalability, reproducibility and dynamic reporting.
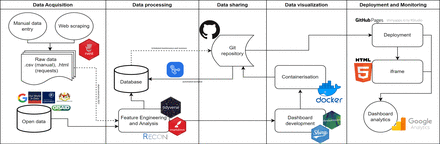
Methods A five-tiered workflow of data acquisition; processing; databasing, sharing, version control; visualisation; and monitoring was used. COVID-19 data were initially collated from press releases and then transitioned to official sources.
Results Key COVID-19 indicators were tabulated and visualised, deployed using open-source hosting in October 2022. The system demonstrated high performance, handling extensive data volumes, with a 92.5% user conversion rate, evidencing its value and adaptability.
Conclusion This cost-effective, scalable solution aids health specialists and authorities in tracking disease burden, particularly in low-resource settings. Such innovations are critical in health crises like COVID-19 and adaptable to diverse health scenarios.
Introduction
The transmission of COVID-19 has led to more than 535 million cases and 6 million deaths in almost every country in the world between January 2020 and June 2022.1 Data and digital solutions are essential in strengthening pandemic management and preparedness.2 3 Dynamic reporting systems have been used as a data solution for decision-making within various fields.4 5 Dynamic reporting systems as part of surveillance systems can be critical for disease control, resource allocation and health planning.6 7 However, developing such a system within lower-income and middle-income countries can be challenging due to the scarcity of human and economic resources.8 Malaysia has experienced high levels of COVID-19 transmission.9 Despite this, there has been a paucity of data collation and translation into epidemiological intelligence for public health interventions within the public space. We aimed to develop an analytical platform that had: (1) a data collation system, (2) an automated analytical and reporting platform, and (3) a system to monitor and evaluate the solution. The solution had to be low-cost and rapidly deployable without programming expertise.
Results
The architecture, developed in October 2020, initially provided semiautomated updates on COVID-19 data and analytics. With Malaysia’s transition into endemicity in April 2022, the dashboard underwent enhancements, deploying extended functionalities in June 2022. This fully automated version collated and published analytics on transmission, disease burden, testing, healthcare capacity, variants and human mobility, using a variety of visual aids (online supplemental appendix 2). Using a web-based plugin, usage statistics were monitored, yielding weekly and biannual performance reports. Remarkably, 92.5% of new users converted into active users, with changes in user engagement paralleling shifts in measures for uptake and transmission dynamics (online supplemental appendix 3). The dashboard drew a median of 0.37% of all traffic to the department website, peaking at 1.9% in November 2020 (online supplemental appendix 4).
Discussion
The escalating COVID-19 transmission within Malaysia necessitated the development of a local, low-cost, rapidly scalable and reproducible solution for data collation, analysis and dashboarding. This initiative was a collaboration of public health experts which yielded a straightforward, agile architecture that can be deployed with limited programming expertise. This open-source model demonstrated robust performance, handling large volumes of data, converting 92.5% of new users into active ones and reflecting stakeholder feedback in its ongoing enhancements. Compared with other open-source solutions in different countries, development by non-expert programmers was the key differentiator, offering potential for use in other low-resource settings. Smaller disease control programmes can use this workflow in developing dynamic reporting systems in said programmes.19 20
Several limitations were observed, including data acquisition challenges, relatively low engagement, limited monitoring of data volumes and collection efficiency, constraints in user experience. Lessons learnt from this project emphasised the importance of stakeholder involvement in the development process, an effective engagement strategy prior to deployment and the necessity of flexibility in the face of changing data formats.
Future iterations should incorporate these insights, and we propose four potential solutions to mitigate current limitations. First, in response to the volatile data sources, improving the system’s adaptability to various data formats is essential. Second, to boost engagement, an inclusive and robust communication strategy must be crafted. Third, detailed data volumes and exact efficiency metrics should also be monitored for a more comprehensive understanding of the system’s impact. Lastly, the user interface can be refined to enhance user experience and stability, without compromising speed and simplicity. Several of these issues were partially addressed in the recent dashboard update of the MOH’s recent use of the GitHub platform for data dissemination and extend functionalities to include more depth in the exploration of indicators.
The value of agile, open-source solutions like ours becomes apparent in combating diseases like COVID-19. The tool’s adaptability makes it beneficial for tracking various health concerns, underscoring the importance of continued development and enhancement.
Conclusion
The development of a low-cost, open-source, rapidly scalable and reproducible workflow as described here can be useful in many situations, especially in low-resource settings. Innovative and low-cost solutions can be critical in crises such as COVID-19 and other health- and healthcare-related scenarios, especially in automating collation, processing and reporting of data. There remains a need for more collaboration in strengthening workflows such as this to allow more rapid adoption of dynamic reporting systems in the health sector.